Welcome to the Drew Schwartz lab
Our goal is to deliver personalized gut microbiome-based risk assessment and antibiotic stewardship for pediatric sepsis.
Bacterial infections are a leading cause of infant death worldwide. Preterm infants hospitalized in neonatal intensive care units (NICU) are the most susceptible to bacterial infections because of underdeveloped skin and gut barriers and other immature organ systems. Because bacterial infection affects up to 40% of preterm infants, antibiotics are frequently given for suspected or confirmed infection. When infants show signs of infection (sepsis), their blood is cultured to look for bacteria, and they are started on antibiotics. While awaiting results, infants are treated with antibiotics targeted at common causes of bloodstream infection, but this approach has negative consequences:
- Many bacteria are becoming increasingly resistant to being killed by the chosen antibiotics.
- When antibiotics are given unnecessarily when no bacteria grow in culture, antibiotics are associated with increased likelihood of death.
- Antibiotics disrupt the normal development of the infant gut microbiome and immune system and increase the abundance of antibiotic-resistant bacteria and risk of further episodes of sepsis.
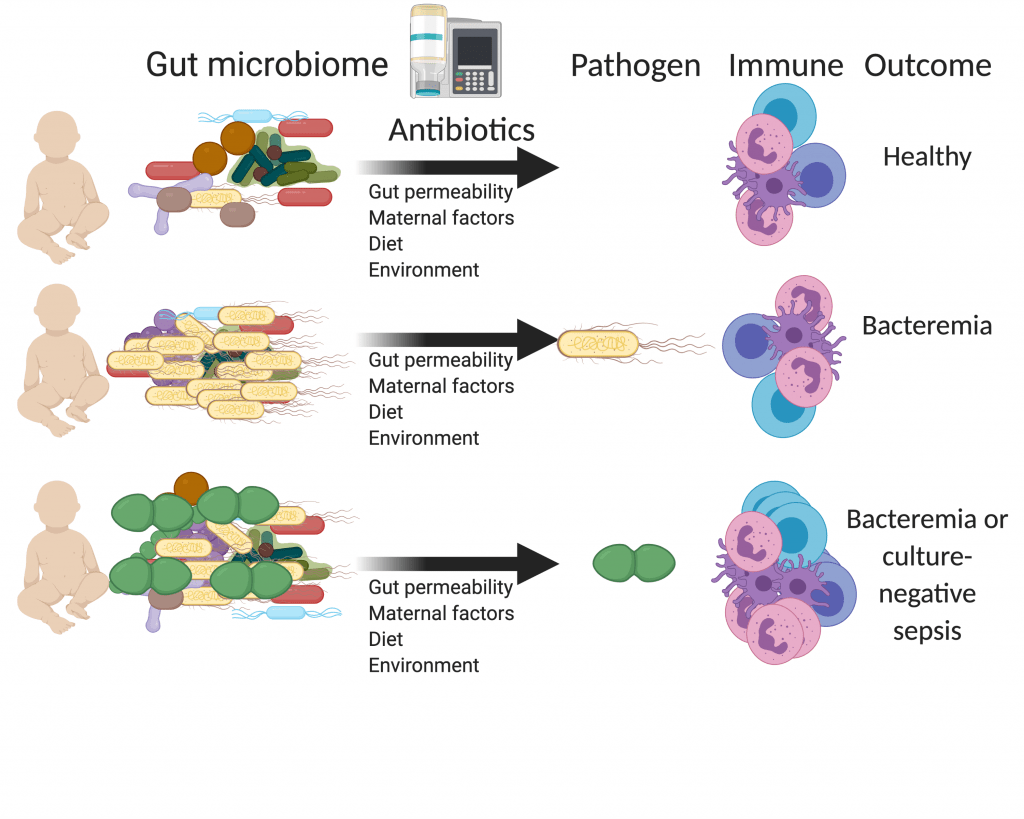
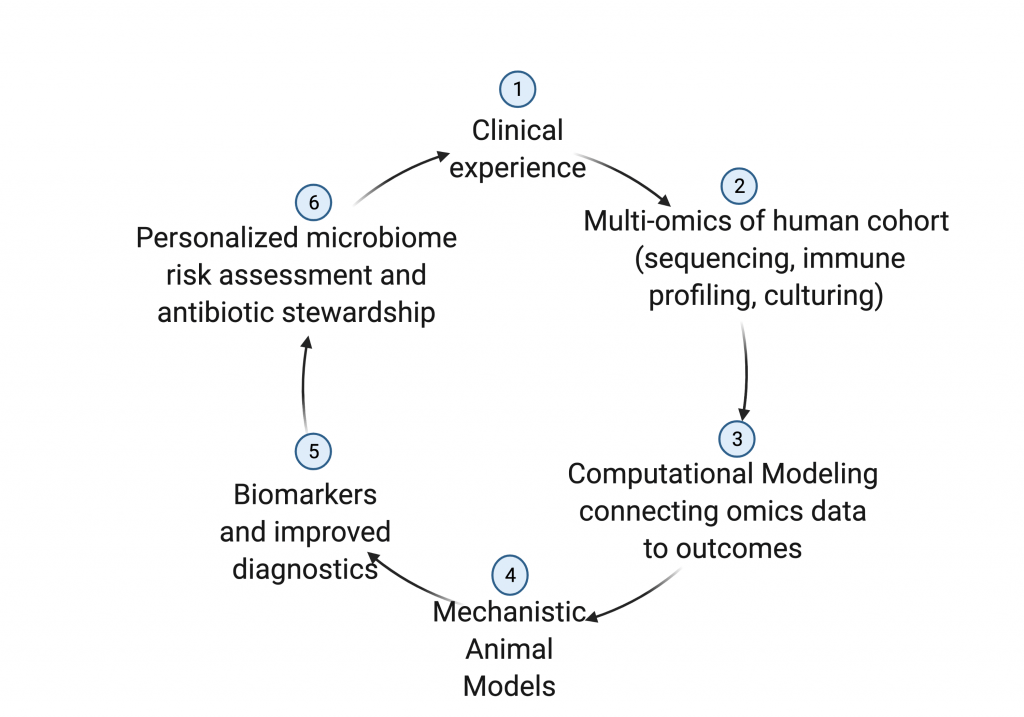
Our lab aims to understand the microbiome-host interface preceding sepsis to lead to better diagnostics and treatments. We use a combination of microbial sequencing and culturing, immune profiling and computational modeling of clinical cohorts and gnotobiotic mouse models to produce algorithms that predict sepsis risk and optimal antibiotic therapies. We hope to translate these findings to clinical populations to reduce morbidity and mortality from serious bacterial infections in pediatric populations.
Learn more about our research…



