
For custom code of published and in-progress manuscripts, please visit our repos on GitHub:
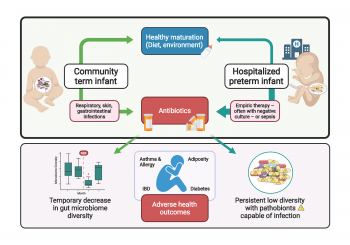
Ten percent of U.S. births are preterm with up to 17% mortality. Up to 35% of preterm infants suffer at least one episode of sepsis, which can be early onset (EOS, first 72 hours after birth) or late onset (LOS, >72 hours after birth). Preterm neonates are treated with antibiotics for culture-negative or culture-positive sepsis which perturb the undeveloped gut microbiome (dominated at this stage by often antibiotic-resistant pathobionts in family Enterobacteriaceae as well as staphylococci and enterococci, impair immune development, enrich for antibiotic resistance, and increase risk of subsequent LOS. We utilize sequencing, immune profiling and computational analyses of clinical cohorts to identify predictors and risk factors of bacteremic sepsis in preterm infants.
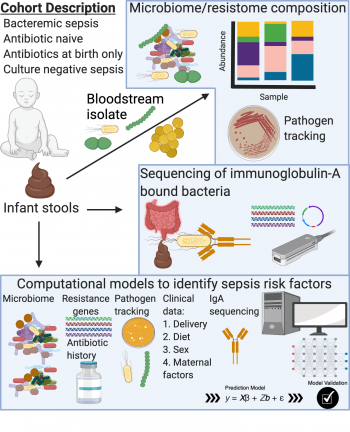
The majority of antibiotic usage in the neonatal intensive care unit is for culture-negative sepsis, where no causative organism is identified. When given for culture-negative sepsis, antibiotic use rate correlates with mortality. We are in desperate need of better diagnostics to identify which infants need antibiotics and which do not. We utilize gut microbiome sequencing, culturing and immune profiling, combined with computational analyses of human clinical cohorts to better understand culture-negative sepsis.
Up to 10% of febrile term infants will experience a serious bacterial infection (urinary tract infection, bacteremia, meningitis) in the first 60 days of life. While awaiting diagnostics in this population, term infants are admitted and treated with broad-spectrum antibiotics. Our lab is trying to develop better predictors of which febrile term infants have a serious bacterial infection versus others. Our goal is to provide better antibiotic stewardship to treat serious bacterial infection and spare antibiotics and their negative consequences more quickly for those without infection. We use computational analyses on gut microbiomes and bacterial isolates from infants with serious bacterial infection and paired controls to better predict risk and deliver appropriate antibiotics.
*These authors contributed equally.
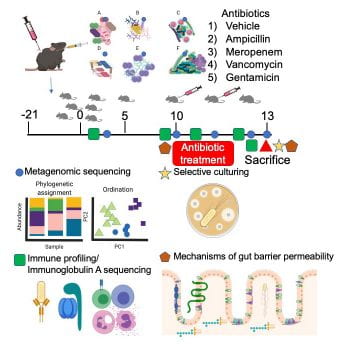
We have developed a neonatal microbiome-humanized mouse model to interrogate the gut microbiome, host immune, and epithelial mechanisms of bacteremic and culture-negative sepsis. We colonize germ-free dams with neonatal microbes (stool or culture collections). Pups are born microbiota-humanized and are treated with pharmacokinetically appropriate doses of subcutaneous antibiotics. Depending on the microbiome-antibiotic combination mice live or die with culture-negative or bacteremic sepsis. This model allows us to probe the gut microbiome-host immune interface to understand how the immune system develops in the context of an individual’s microbiome.
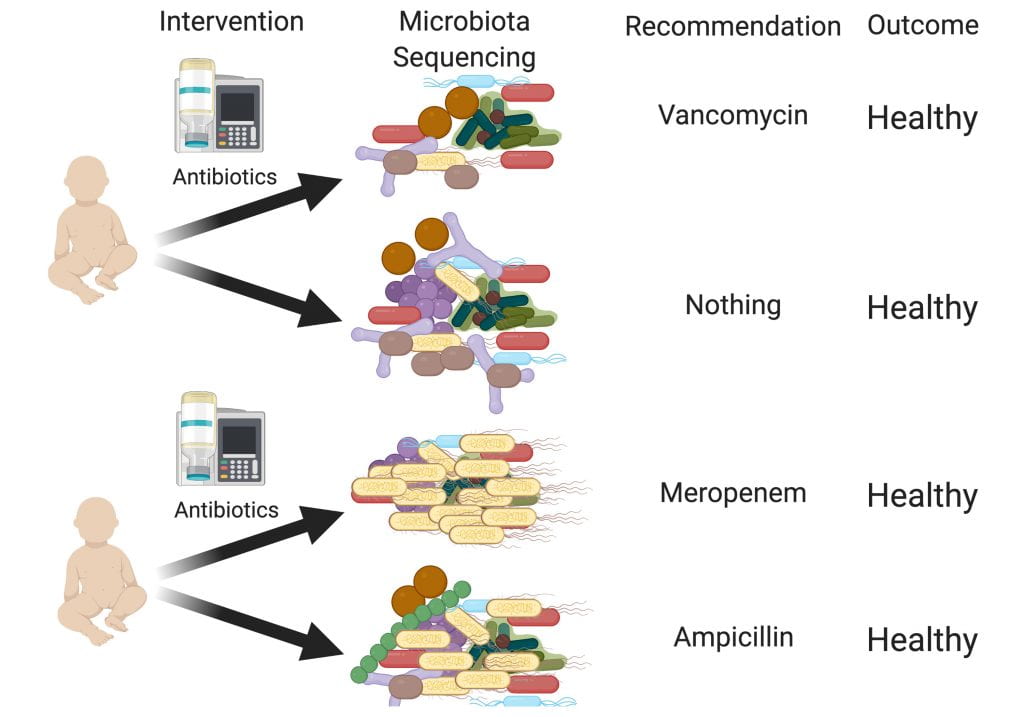
We will perform clinical trials to determine the impact of personalized microbiome-based risk assessment and antibiotic stewardship on the gut microbiome and health of preterm infants. Our goal is to prevent sepsis and minimize damage from necessary and unnecessary antimicrobials.
With generous support …
Work in the Schwartz lab is supported by an NIH K08 from NIAID, a Doris Duke Charitable Foundation Physician Scientist Fellowship, and a Thrasher Research Fund Early Career Award.
We acknowledge past support from a Pediatric Infectious Disease Society and St. Jude Children’s Hospital Fellowship in Basic and Translational Research.